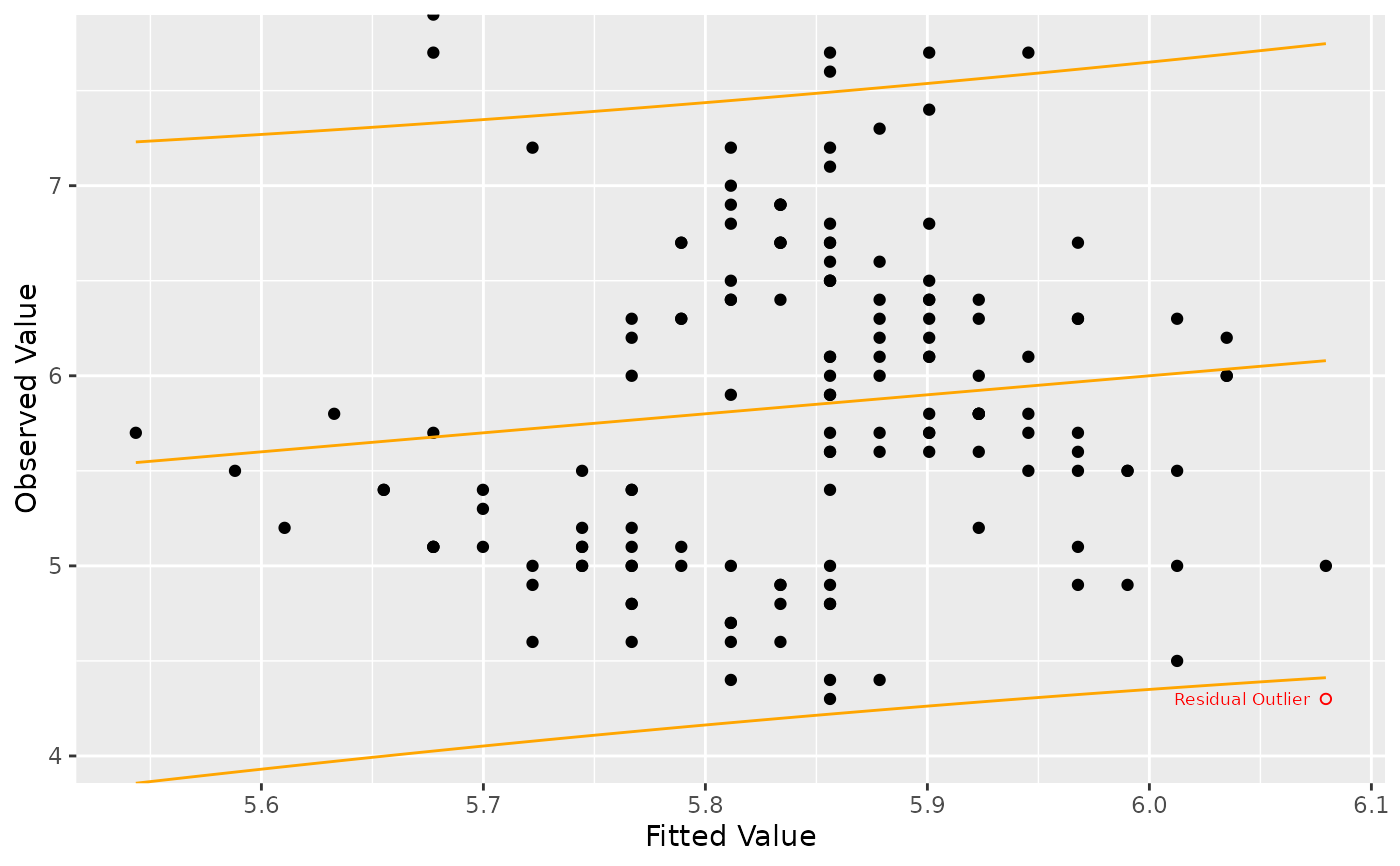
Generates a scatter plot of fitted values versus observed values from a linear model, with identification of outlier points and optional prediction interval. The plot includes a reference line y = x for assessing linearity.
Usage
lm_plot.fit(
mdl,
...,
pred.intvl = TRUE,
parms = lm_plot.parms(mdl),
df = lm_plot.df(mdl, parms = parms)
)Arguments
- mdl
A fitted model object (typically from
lm).- ...
Additional arguments (not currently used).
- pred.intvl
(logical, default = TRUE) Option to show prediction interval bounds of fitted values.
- parms
List of plotting parameters, usually from
lm_plot.parms().- df
Data frame with augmented model data. Defaults to
lm_plot.df(mdl).
Value
A ggplot object representing the fitted vs. observed plot. Included as an attribute "parm" is a list containing:
limPlotted limits onxandyaxes,pred.intvlOption to show prediction interval,
Details
The plot visualizes fitted versus observed values, includes a diagonal reference line, marks outliers, and can optionally display prediction intervals. Outlier and regular points can be labeled. This plot is useful for visually assessing linearity and model fit quality.
Examples
mdl <- lm(Sepal.Length ~ Sepal.Width, data = iris)
lm_plot.fit(mdl)