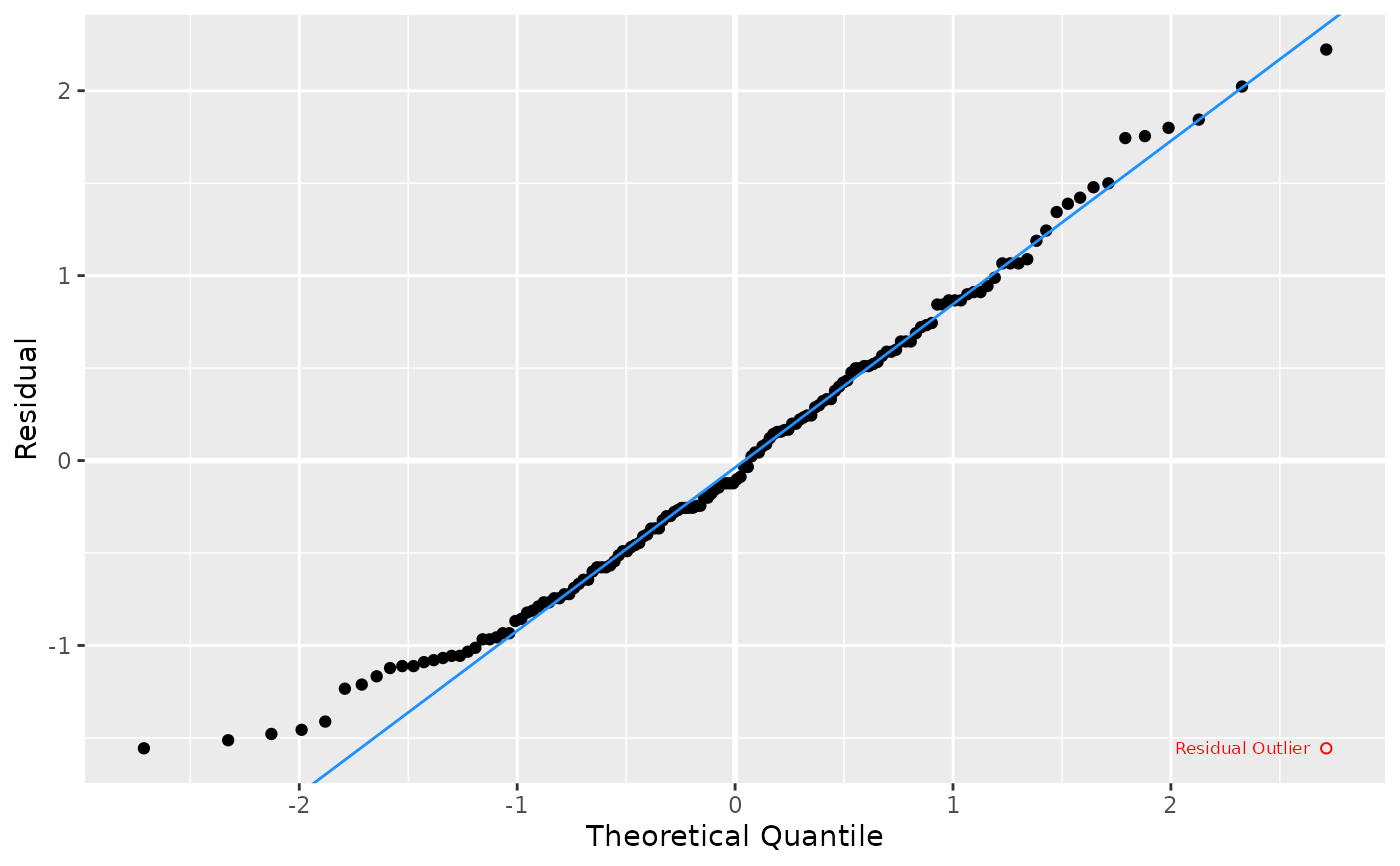
Produces a Q-Q plot of residuals from a linear model to test for normality, optionally annotating outlier points and adding the Shapiro-Wilk test p-value.
Usage
lm_plot.qq(
mdl,
...,
pval.SW = FALSE,
parms = lm_plot.parms(mdl),
df = lm_plot.df(mdl, parms = parms)
)Arguments
- mdl
A fitted model object (typically from
lm).- ...
Additional arguments (not currently used).
- pval.SW
(logical, default = FALSE) indicates whether to include Shapiro-Wilk p-value on the plot.
- parms
List of plotting parameters, usually from
lm_plot.parms().- df
Data frame with augmented model data. Defaults to
lm_plot.df(mdl).
Value
A ggplot object representing the quantile-quantile plot. Included as an attribute "parm" is a list containing:
limPlotted limits onxandyaxes,pval.SWOption to show Shapiro-Wilk p-value,DWThehtestobject with Shapiro-Wilk test results.
Details
The plot visualizes the distribution of residuals against theoretical normal quantiles. Points are colored and shaped by outlier status, and a reference Q-Q line is added. Optionally, outlier and regular points can be labeled. If enabled, the Shapiro-Wilk normality test p-value is displayed.
Examples
mdl <- lm(Sepal.Length ~ Sepal.Width, data = iris)
lm_plot.qq(mdl)